forked from ac812/reproducibility-training
-
Notifications
You must be signed in to change notification settings - Fork 6
/
Copy pathrmarkdown.Rmd
727 lines (515 loc) · 28.3 KB
/
rmarkdown.Rmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209
210
211
212
213
214
215
216
217
218
219
220
221
222
223
224
225
226
227
228
229
230
231
232
233
234
235
236
237
238
239
240
241
242
243
244
245
246
247
248
249
250
251
252
253
254
255
256
257
258
259
260
261
262
263
264
265
266
267
268
269
270
271
272
273
274
275
276
277
278
279
280
281
282
283
284
285
286
287
288
289
290
291
292
293
294
295
296
297
298
299
300
301
302
303
304
305
306
307
308
309
310
311
312
313
314
315
316
317
318
319
320
321
322
323
324
325
326
327
328
329
330
331
332
333
334
335
336
337
338
339
340
341
342
343
344
345
346
347
348
349
350
351
352
353
354
355
356
357
358
359
360
361
362
363
364
365
366
367
368
369
370
371
372
373
374
375
376
377
378
379
380
381
382
383
384
385
386
387
388
389
390
391
392
393
394
395
396
397
398
399
400
401
402
403
404
405
406
407
408
409
410
411
412
413
414
415
416
417
418
419
420
421
422
423
424
425
426
427
428
429
430
431
432
433
434
435
436
437
438
439
440
441
442
443
444
445
446
447
448
449
450
451
452
453
454
455
456
457
458
459
460
461
462
463
464
465
466
467
468
469
470
471
472
473
474
475
476
477
478
479
480
481
482
483
484
485
486
487
488
489
490
491
492
493
494
495
496
497
498
499
500
501
502
503
504
505
506
507
508
509
510
511
512
513
514
515
516
517
518
519
520
521
522
523
524
525
526
527
528
529
530
531
532
533
534
535
536
537
538
539
540
541
542
543
544
545
546
547
548
549
550
551
552
553
554
555
556
557
558
559
560
561
562
563
564
565
566
567
568
569
570
571
572
573
574
575
576
577
578
579
580
581
582
583
584
585
586
587
588
589
590
591
592
593
594
595
596
597
598
599
600
601
602
603
604
605
606
607
608
609
610
611
612
613
614
615
616
617
618
619
620
621
622
623
624
625
626
627
628
629
630
631
632
633
634
635
636
637
638
639
640
641
642
643
644
645
646
647
648
649
650
651
652
653
654
655
656
657
658
659
660
661
662
663
664
665
666
667
668
669
670
671
672
673
674
675
676
677
678
679
680
681
682
683
684
685
686
687
688
689
690
691
692
693
694
695
696
697
698
699
700
701
702
703
704
705
706
707
708
709
710
711
712
713
714
715
716
717
718
719
720
721
722
723
724
725
726
727
---
title: "R Markdown"
author: "Alexia Cardona"
bibliography: references.bib
link-citations: yes
output:
html_document:
highlight: pygments
toc: true
toc_float:
collapsed: true
toc_depth: 6
---
<br/>
# RStudio
We will be using RStudio both for this R Markdown session and also during the GitHub session. RStudio is currently a very popular Integrated Development Environment (IDE) for working with R. An IDE is an application used by software developers that faciliates programming by offering source code editing, building and debugging tools all integarated into one application. To function correctly, RStudio needs R and therefore both need to be installed on your computer.
The RStudio Desktop open-source product is free under the
[Affero General Public License (AGPL) v3](https://www.gnu.org/licenses/agpl-3.0.en.html). [Other versions of RStudio](https://www.rstudio.com/products/rstudio/) are also available.
We will use RStudio IDE to write code, navigate the files on our computer,
inspect the variables we are going to create, and visualize the plots, tables and the notebooks that we will
generate. RStudio can also be used for version control and we will be looking at this later on.

RStudio is divided into 4 "Panes": the **Source** for your scripts and documents
(top-left, in the default layout), your **Environment/History/Build/Git** (top-right),
your **Files/Plots/Packages/Help/Viewer** (bottom-right), and
the R **Console** (bottom-left). The placement of these
panes and their content can be customized (see menu, Tools -> Global Options ->
Pane Layout).
One of the advantages of using RStudio is that all the information
you need to write code is available in a single window. Additionally, with many
shortcuts, autocompletion, and highlighting for the major file types you use
while developing in R, RStudio will make typing easier and less error-prone.
\
# Creating an R Markdown project
Before starting to write code in RStudio, we need to create an R Project. The idea behind an R project is to have a workspace where you can keep all the files and settings associated with the project together. In that way, next time you open the R Project it would be easier to resume work. To create an R notebook project for the web in RStudio follow the following steps:
1. `File` -> `New Project` -> `New Directory` -> `Simple Rmarkdown website`.
2. Specify where you would like to create your project. This would be your **working directory**.
3. Click `Create Project`.
RStudio automatically generates a template for you to develop further. To see how this template would look as a website, click on the `Build` tab on the top right hand side pane of RStudio and then click `Build Website`.
<img src="img/build-website.png" width="400"/>
RStudio's default preferences generally work well, but saving a workspace to
`.RData` can be cumbersome, especially if you are working with larger datasets as this would save all the data that is loaded into R into the `.RData` file.
To turn that off, go to `Tools` -> `Global Options` and select the 'Never' option
for `Save workspace to .RData' on exit.`

\
## Working directory
Whenever we are working on a project in RStudio, it is good practice to keep a set of related files *e.g.,* data, images, and code, self-contained in a single folder, called the **working directory**. All of the scripts within this folder can then use **relative paths** to files in the working directory that indicate where inside the project a file is located (as opposed to **absolute paths**, which point to where a file is on a specific computer). Working this way makes it
a lot easier to move your project around on your computer and share it with others without worrying about whether or not the underlying scripts will still work.
> **Absolute vs Relative paths examples**
>
> **Relative path**: data/dataset1.txt
>
> **Absolute path**: C:/Users/User1/Documents/R/my-first-project/**data/dataset1.txt**
Using RStudio projects makes it easy to organise your files in the project and ensures that your working directory is set properly. RStudio shows your current working directory at the top of your window:

Another way to check your working directory is by typing `getwd()` in the console pane. If for some
reason your working directory is not what it should be, you can change it in the
RStudio interface by navigating in the file browser where your working directory
should be, and clicking on the blue gear icon `More`, and select `Set As Working
Directory`. Alternatively you can type `setwd("/path/to/working/directory")` in the console to
reset your working directory (not recommended). However, your scripts should not include this line
because it will fail on someone else's computer.
Using a consistent folder structure across your projects will help keep things
organized, and will also make it easy to find things in the future. This
can be especially helpful when you have multiple projects. In general, you may
create directories (folders) for **data**, and **images**.
- **`data/`** Use this folder to store your raw data and intermediate
datasets you may create for the need of a particular analysis. For the sake
of transparency and [provenance](https://en.wikipedia.org/wiki/Provenance),
you should *always* keep a copy of your raw data accessible and do as much
of your data cleanup and preprocessing programmatically (*i.e.,* with scripts,
rather than manually). Separating raw data from processed data
is also a good idea. For example, you could have files
`data/raw/survey.plot1.txt` and `data/raw/survey.plot2.txt` kept separate from
a `data_output/survey.csv` file generated by the
`scripts/01.preprocess.survey.R` script.
- **`img/`** This would be a place to keep figures that you would like to display in your notebook.
- **`scripts/`** This would be the location to keep your R scripts for
different analyses or plotting.
You may want additional directories or subdirectories depending on your project
needs, but these should form the backbone of your working directory.
The working directory is an important concept to understand. It is the place
from where R will be looking for and saving the files. When you write code for
your project, it should refer to files in relation to the root of your working
directory and only need files within this structure.
\
# R Markdown
An R Markdown file is made up of 3 basic components:
- header
- markdown
- R code chunks
In this course we will assume that the output of our report is an .html file. HTML files are files for web pages. This means that the report that we will generate can be easily deployed on the web.
## Header
The markdown document starts with an optional header in YAML format known as the YAML metadata. In the example below the title, author and date are specified in the header. Other options can be specified in the header such as table of content which we will look at later on in the course.
```
---
title: "My first notebook"
author: Alexia Cardona
date: 18 February 2020
---
```
> **Hint**
>
> Press the <img src="img/knit.png" width="90"/> button to see how the report will look like after you make changes to the .Rmd file.
\
## Markdown {#markdown-header}
The text following the header in an R Markdown file is in Markdown syntax. This is the syntax that gets converted to HTML format once we click on the Knit button or the Build website button. The philosophy behind Markdown is that it should be easy to write and easy to read.
The full documentation of the Markdown syntax can be found at https://pandoc.org/MANUAL.html. However this can be a bit difficult to follow for beginners, therefore below is a simplified version of the Markdown syntax.
\
### Headings
Below is the `Markdown code` you need to use to specify headings at different levels and the rendered output respectively below the code:
`# Heading 1`
<p style="font-weight:500; font-size:34px">Heading 1</p>
`## Heading 2`
<p style="font-weight:500; font-size:30px">Heading 2</p>
`### Heading 3`
<p style="font-weight:500; font-size:24px">Heading 3</p>
`#### Heading 4`
<p style="font-weight:500; font-size:18px">Heading 4</p>
\
### Inline text formatting
To make text **bold** use `**double asterisks**` or `__double underscores__`.
To make text *italic* use `*asterisks*` or `_underscores_`.
To make text ^superscript^ use `^caret^`.
To make text ~subscript~ use `~tilde~`.
To mark text as `inline code` use `` `backticks` ``.
To ~~strikethrough~~ text use `~~double tilde~~`.
\
### Line breaks
To create a line break, put more than 2 spaces at the end of a sentence or place `\` in a new line followed by a new line <img src="img/enter.png" width="20"/>.
\
>### Challenge
>Replicate the output of the text below in R Markdown.
>The text comes from [a paper by Monya Baker](https://www.nature.com/news/1-500-scientists-lift-the-lid-on-reproducibility-1.19970) that was published in 2016 in the Nature journal that triggered the discussion about Reproducibility Crisis.

> <details>
> <summary>Show Answer</summary>
>```
>---
>title: "1,500 scientists lift the lid on reproducibility"
>author: Monya Baker
>date: 25 May 2016
>---
>\
>
>## Survey sheds light on the 'crisis' rocking research.
>
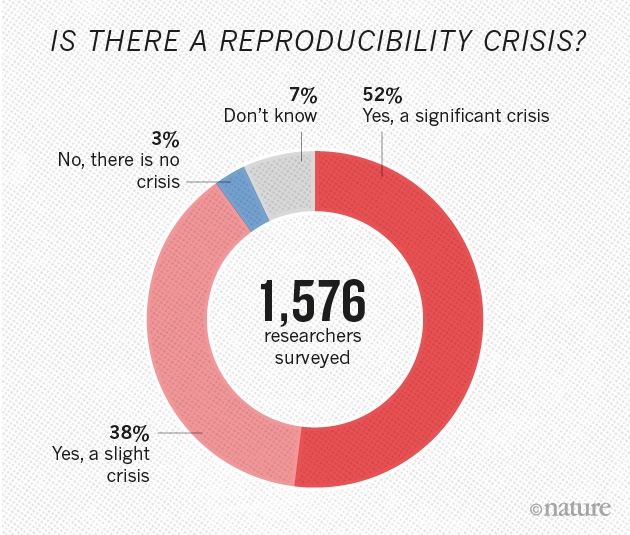
>**More than 70% of researchers have tried and failed to reproduce another scientist's experiments**, and more than half have failed to reproduce their own experiments. Those are some of the telling figures that emerged from *Nature*'s survey of 1,576 researchers who took a brief online questionnaire on reproducibility in research.
>
>The data reveal sometimes-contradictory attitudes towards reproducibility. Although 52% of those surveyed agree that there is a significant 'crisis' of reproducibility, less than 31% think that failure to reproduce published results means that the result is probably wrong, and most say that they still trust the published literature.
>```
> </details>
>
\
### Links
#### Linking text to Headers
To link text to a header you would need to specify an identification tag next to a header as follows:
`# Markdown {#markdown-header}`
Then to link text to this header use `[link to header](#markdown-header)`. This will be rendered as [link to header](#markdown-header).
\
#### Linking text to a webpage
To create a link to a webpage use `[text of link](https://training.cam.ac.uk/bioinformatics/event-timetable)`. This is rendered as [text of link](https://training.cam.ac.uk/bioinformatics/event-timetable).
\
#### Footnotes
To indicate a footnote use`[^1]` and, for example, indicate another one as`[^2]` and then specify the wordings of the footnotes as:
`[^1]: This is the first footnote.`
`[^2]: This is the second footnote.`
You do not need to put footnotes at the end of the document for them to be rendered there. This example is rendered as follows:
To indicate a footnote use[^1] and, for example, indicate another one as[^2] and then specify the wordings of the footnotes as:
[^1]: This is the first footnote.
[^2]: This is the second footnote.
\
### Lists
#### Ordered lists
To create an ordered list use the following syntax:
```
1. Item 1
2. Item 2
3. Item 3
```
This is rendered as:
1. Item 1
2. Item 2
3. Item 3
Use 4 spaces to indent an item if you would like to have sub-lists:
```
1. Item 1
2. Item 2
3. Item 3
a. Item 3a
i. Item 3ai
ii. Item 3aii
b. Item 3b
c. Item 3c
4. Item 4
```
1. Item 1
2. Item 2
3. Item 3
a. Item 3a
i. Item 3ai
ii. Item 3aii
b. Item 3b
c. Item 3c
4. Item 4
\
#### Unordered lists
In an unordered bulletted list, each item begins with `*`, `+` or `-`. Example:
```
* Item 1
* Item 2
* Item 3
* Item 3a
* Item 3ai
* Item 3aii
* Item 3b
* Item 3c
* Item 4
```
Will be rendered as:
* Item 1
* Item 2
* Item 3
* Item 3a
* Item 3ai
* Item 3aii
* Item 3b
* Item 3c
* Item 4
\
#### Tasks list
Tasks list can be done using the following syntax:
```
- [ ] an unchecked task list item
- [x] checked item
```
This will be rendered as:
- [ ] an unchecked task list item
- [x] checked item
\
### Inserting images
To insert an image use the following syntax: ``. Example:
``
Will be rendered as:

\
>### Challenge
>Continue working on the example we created in Challenge 1 to extend it to an output that is similar to [this](example2.html).
> <details>
> <summary>Show Answer</summary>
>```
>---
>title: "1,500 scientists lift the lid on reproducibility"
>author: Monya Baker
>date: 25 May 2016
>---
>\
>
>## Reproducibilty Crisis
>
>**More than 70% of researchers have tried and failed to reproduce another scientist's experiments**, and more than half have failed to reproduce their own experiments. Those are some of the telling figures that emerged from *Nature*'s survey of 1,576 researchers who took a brief online questionnaire on reproducibility in research.
>
>The data reveal sometimes-contradictory attitudes towards reproducibility. Although 52% of those surveyed agree that there is a significant 'crisis' of reproducibility, less than 31% think that failure to reproduce published results means that the result is probably wrong, and most say that they still trust the published literature.
>
>
>
>\
>
>### The Survey
>
>In the survey [^1] respondents were asked to rate 11 different approaches to improving reproducibilty in science. Below is the list order by the most highly rated:
>
>* Better understanding of statistics
>* Better mentoring/supervision
>* More robust design
>* Better teaching
>* More within-lab validation
>* Incentives for better practice
>* Incentives for formal reproduction
>* More external-lab validation
>* More time for mentoring
>* Journals enforcing standards
>* More time checking notebooks
>
>[^1]: The survey can be downloaded [here](https://www.nature.com/news/polopoly_fs/7.36741!/file/Reproduciblility%20Questionnaire.doc).
>
>```
> </details>
>
\
### Tables {#tables}
Use `|`and `-` to create a table as follows:
```
| Column 1 | Column 2 |
| ----------- | ----------- |
| Item 1,1 | Item 1,2 |
| Item 2,1 | Item 2,2 |
```
This is rendered as:
| Column 1 | Column 2 |
| ----------- | ----------- |
| Item 1,1 | Item 1,2 |
| Item 2,1 | Item 2,2 |
Table alignments can be done using the following syntax:
```
| Left align | Center align | Right align |
| :--- | :----: | ---: |
| Item 1,1 | Item 1,2 | Item 1,3 |
| Item 2,1 | Item 2,2 | Item 2,3 |
```
This is rendered as:
| Left align | Center align | Right align |
| :--- | :----: | ---: |
| Item 1,1 | Item 1,2 | Item 1,3 |
| Item 2,1 | Item 2,2 | Item 2,3 |
\
### Blocks
Blocks in the notebook can be created by using the `>` sign as follows:
`> Example of a block`
This is rendered as:
> Example of a block
If you would like to add **code blocks**, use ` ``` ` before and after the code as follows:
~~~
```
print("Hello world")
x <- 1+2
print(x)
```
~~~
This will be rendered as:
```
print("Hello world")
x <- 1+2
print(x)
```
\
### Adding a table of contents
To add a table of contents to your report add the following to the YAML header:
```
output:
html_document:
toc: true
toc_float: true
```
By default all headings up to level 3 headings are displayed in the table of contents. You can adjust this by specifying `toc_depth` as following:
```
output:
html_document:
toc: true
toc_depth: 4
toc_float: true
```
\
### (Optional) Adding references
Adding references and citations in Markdown is not as easy as reference manager software such as Mendeley. To be able to create citations you will need to create a bibliography file with all the references in it. Here is an [example of a bibliography file](references.bib). The bibligraphy file has to be placed in the same folder as the one where the `.Rmd` file is. Next, add the following to the YAML header:
```
bibliography: references.bib
link-citations: yes
```
Your YAML header should now look like:
```
---
title: "My first notebook"
author: Alexia Cardona
date: 18 February 2020
bibliography: references.bib
link-citations: yes
---
```
To cite a reference use the `@` together with the ID of the reference. Example:
`Citation to my paper @cardona2014 and @cardona2019`
Will be rendered as:
Citation to my paper @cardona2014 and @cardona2019
To add the bibliography at the end of the report add a References heading at the end of the report:
`# References`
See [references](#refs) at the end of the report.
See https://rmarkdown.rstudio.com/authoring_bibliographies_and_citations.html for more information.
\
## Inserting R Code in Markdown
So far we have not used any R code and all the code we used so far is in Markdown. As the name suggests, R Markdown files contain Markdown and R. R Markdown files have a `.Rmd` extension. Using R in a Markdown document is useful if we want dynamically generated information such as integrating our analysis in our report. Some operations that we might need to do are; loading our dataset, performing some operations on the dataset and displaying results, either in a table or in a plot. We will be exploring how we can integrate R code in R Markdown in the following worked example.
>1. Create a new R Markdown document (`File` -> `New File` -> `R Markdown` )
>2. The file created should already contain some R Markdown code and it should look like this:
>
>3. Delete the existing code leaving only the R Markdown header so that we can start building our code as we go over the worked example.
>4. Name your R Markdown document as RNotebookExample.Rmd
\
### The dataset
Our worked example uses data from [gapminder](https://www.gapminder.org/). Let us download the `gapminder_data.csv` dataset into our project. Create a `data` folder in your working directory. Download the dataset into the `data` folder as follows:
```
download.file(url="https://raw.githubusercontent.com/cambiotraining/reproducibility-training/master/data/gapminder_data.csv", destfile="data/gapminder_data.csv")
```
The `gapminder_data.csv` dataset that is loaded into the notebook contains the life expectancy and GDP per capita data for each country per year from 1952 to 2007. The dataset has the following columns:
| Column | Description |
|------------------|-----------------------------------------------------------|
| country | Country |
| year | Year the life expectency and GDP per capita index applies |
| pop | Population size |
| continent | Continent |
| lifeExp | Life expectancy |
| gdpPercap | GDP per capita index |
The dataset contains data for different countries in the world. For our analysis we would like to look at only the European countries in the dataset. This means that we would need to manipulate the dataset before we can display the results.
\
### R code chunks
To be able to insert R code inside a notebook you will have to insert it inside an R code chunk to be able to execute it. To do this either click on the `Insert` button at the top of the Source panel in RStudio:
<img src="img/insert-r-chunk.png" width=150/>
This creates an **R code chunk** as follows:

Alternatively, you can type the r code chunk. The R code should be placed in between the triple backticks. Note that on the right hand side of the R code chunk there is a green "play" button that will run the R code chunk if pressed. By default, when the R Markdown document is knitted, the R code will be executed and the R code chunk is displayed before the executed code.
\
### Chunk options {#chunk-options}
As mentioned, by default the chunk output will be displayed exactly after the R code chunk and the R code chunk is also displayed in the rendered page. If you would like to have different settings, you can specify these as arguments in the `{}` part of the R code chunk.
These chunk options include:
* `include = FALSE` do not display the code and results in the page after it is knitted. The R code however still runs and therefore the variables or results in this code chunk can be used by the other chunks.
* `echo = FALSE` does not display the code, but it displays the results in the rendered file.
* `message = FALSE` does not display any messages that are generated by the code chunk in the rendered file.
* `warning = FALSE` does not display warnings that are generated by code chunk in the rendered file.
A full list of options that can be used in the R code chunks is found [here](https://www.rstudio.com/wp-content/uploads/2015/03/rmarkdown-reference.pdf).
\
### Chunk label
Most likely you will specify several R code chunks in your R Markdown file. It is recommended practice to label each R code chunk. This will also help navigating your code easily. At the bottom right of your RStudio you should see the code navigator where you can navigate the different code chunks:

To label your chunk just add a label next to `r` in your R code chunk as follows:
````markdown
`r ''````{r label-name}
```
````
For our worked example, create an R code chunk and name it as `load-data` and add the following code into your first chunk:
````markdown
`r ''````{r load-data}
#load tidyverse library
library(tidyverse) # used for data manipulation
library(rmarkdown) # used for paged_table function
library(kableExtra) # used for table
library(ggpubr) #used for ggarrange function
#read file into R
pop_data <- read_csv("data/gapminder_data.csv")
#create a table with data from European countries in 2007 showing the countries
#with the largest life expectancy at the top
euro_data_tbl <- pop_data %>%
filter(continent == "Europe" & year == 2007) %>%
select(-continent, -year) %>%
arrange(desc(lifeExp)) %>%
rename(Country = country, "Population Size" = pop,
"Life Expectancy" = lifeExp, "GDP" = gdpPercap)
```
````
We will look at some functions present in the `rmarkdown` and `kableExtra` packages later on. Knit the `RNotebookExample.Rmd` file and see what happens. You may notice that both the code and the output that you would expect to be printed in the console when you run the code are printed in the notebook. We would like to have only the code printed in the notebook. To do that we need to specify the `message=FALSE` as the chunk option for our `load-data` chunk as discussed previously in the [chunk options section](#chunk-options).
````markdown
`r ''````{r load-data, message=FALSE}
````
If you knit `RNotebookExample.Rmd` again you will see that now only the code is being printed, which is what we wanted. In the `load-data` R code chunk, we have loaded the packages we will be using in our report and also the data we will be working with. We then filtered our dataset to contain data from only European countries in 2007 ordered by life expectancy. The next step is to display this filtered dataset that is saved in the variable `eur_data_tbl` in a table.
\
### Tables in R Markdown
When you have a large dataset, creating a table manually in R Markdown as we did [here](#tables) is not really feasible. Also if you are really trying to make your research reproducible, then, you will need to generate most of the tables dynamically and therefore you will not be able to manually type them in markdown. This is where R Markdown comes in as there is a way to display tables dynamically. In this course we will look at two ways of displaying tables dynamically, using the functions:
1. `kbl()`
2. `paged_table()`
\
#### `kbl()`
The function `kbl()` is present in the package `kableExtra`, a popular package to display tables in R Markdown. There is extensive documentation on how you can use the functions within the `kableExtra` package to draw tables [here](https://cran.r-project.org/web/packages/kableExtra/vignettes/awesome_table_in_html.html) which has good examples on how to change the different settings of a table, such as font, width, highlights, *etc...*
As a first step, to draw a table you need to use the function `kbl()`. The `kbl()` function takes as an input the dataset you want to display in a table, *e.g., * `kbl(euro_data_tbl)`.
We have already loaded the package `kableExtra` in the `load-data` R code chunk. Create a new R code chunk called `kbl-table` and draw the a table with the `eur_data_tbl` tibble.
````markdown
`r ''````{r kbl}
euro_data_tbl %>%
kbl()
```
````
Let us add a style to the table so that it looks more attractive. The `kable_styling()` function is the function used to customise the way the table looks. We are going to add the `striped` style so that the table have striped rows and also the `hover` style so that when you move your mouse over the table you can see it hover.
````markdown
`r ''````{r kbl}
euro_data_tbl %>%
kbl() %>%
kable_styling(bootstrap_options = c("striped", "hover")) %>%
```
````
\
>##### Challenge
>
>1. Our table is getting prettier. Try a few styles with `kable_styling()`. Look at the [documentation](https://cran.r-project.org/web/packages/kableExtra/vignettes/awesome_table_in_html.html#Table_Styles) for ideas. After you have tried a few styles, set your R Markdown file to look like [this](RNotebookExample1.Rmd)
>
>2. Experiment using the different chunk options and see what the output will be link with each option. Refer to [chunk options section](#chunk-options).
>
\
#### `paged_table()`
If you have a very long table to display, the best way would be to use pagination where the contents of the table are split into multiple tabs. The function `paged_table` in the `rmarkdown` library can be used to do this. It is also very simple to use.
Create a new R code chunk called `paged-table` and use the `paged_table()` function to display the euro_data_tbl table as follows:
````markdown
`r ''````{r paged-table}
paged_table(euro_data_tbl)
```
````
\
### Adding images
Adding images is straightforward and does not require using a specific R Markdown function.
>#### Challenge
>
>Create a new dataset `euro_data_fig` by filtering the `pop_data` tibble to contain only data from Europe. Draw a plot to display the `lifeExp` on the y axis and `year` on the x axis. Use `geom_violin()` to draw this as a violin plot to show the distrubution of the data across each year and save it in a `euro_plot` variable.
>
>\
>
>[Link to Answer](RNotebookExample3.Rmd)
\
### Optional
In reports or publications, sometimes you would need to place two or more figures next to each other. `ggarrage()` is a function present in the `ggpubr` pacakge that can help with this. The `ggarrange()` function takes as an input the list of plots to plot. You can specify the number of columns and rows to plot by the ncol and nrow arguments respectively. Another thing that you might need is a lable for each plot so that you can specify the description in your caption. Use the labels argument to specify labels.
### Challenge
> Create a new dataset `uk_data_fig` by filtering the `pop_data` tibble to contain only data from the United Kingdom. Draw a scatter plot to display the `lifeExp` on the y axis and `year` on the x axis and save it in a `uk_plot` variable.
> Draw the `euro_plot` created in the previous challenge next to a `uk_plot` using the `ggarrange()` function. Label the plots A and B respectively.
>
>\
>[Link to Answer](RNotebookExample4.Rmd)
# Summary
Through R notebooks we can now integrate our R code and our results in one readable document that we can share with our collaborators. In this way the code and the results have become one item. For inspiration, examples of R notebooks can be found at https://rpubs.com/
\
# References {#refs}