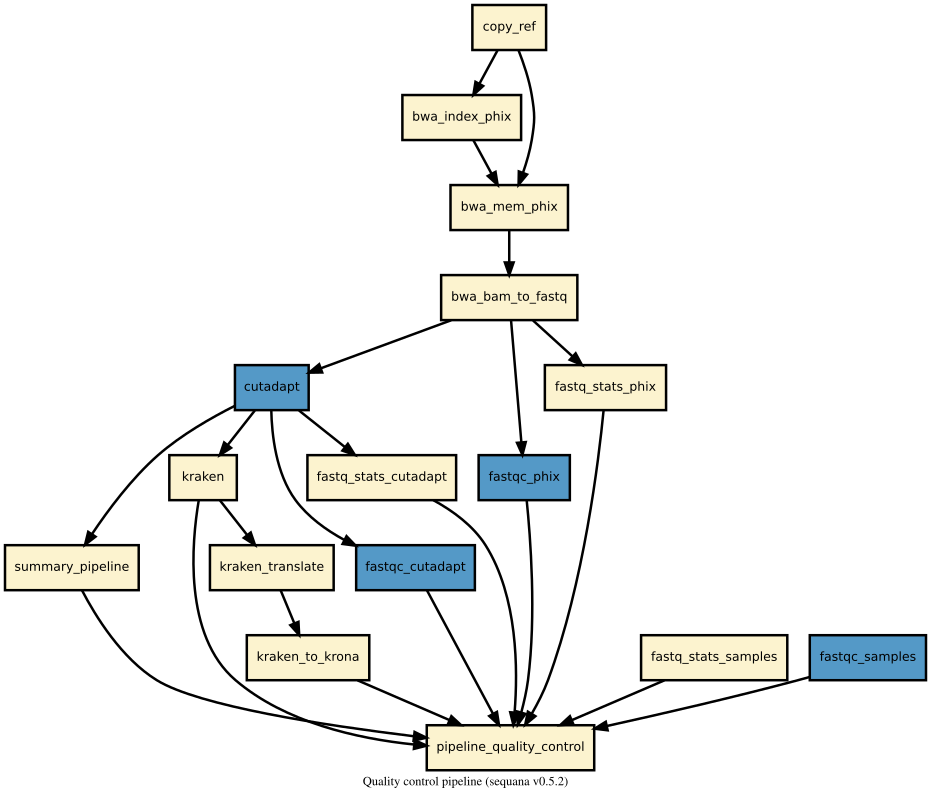
This pipeline is not maintained anymore but should be functional. It is a short-read quality control pipeline from the Sequana project. We would recommend to use the fastqc, demultiplex, and multitax pipelines instead.
| Overview: | A quality control pipeline for illumina data set. This pipeline removes contaminants (e.g. Phix), performs fastqc, adapter cleaning and trimming and checks for contaminants |
|---|---|
| Input: | Raw fastq files |
| Output: | Cleaned fastQ files, remove phix and adapters + taxonomy |
| Status: | production. not maintained. Please use sequana_fastqc and sequana_multitax pipeline instead |
| Citation: | Cokelaer et al, (2017), ‘Sequana’: a Set of Snakemake NGS pipelines, Journal of Open Source Software, 2(16), 352, JOSS DOI doi:10.21105/joss.00352 |
You must install Sequana first:
pip install sequana
Then, just install this package:
pip install sequana_quality_control
sequana_quality_control --help sequana_quality_control --input-directory DATAPATH
This creates a directory with the pipeline and configuration file. You will then need to execute the pipeline:
cd quality_control sh quality_control.sh # for a local run
This launch a snakemake pipeline. If you are familiar with snakemake, you can retrieve the pipeline itself and its configuration files and then execute the pipeline yourself with specific parameters:
snakemake -s quality_control.rules -c config.yaml --cores 4 --stats stats.txt
Or use sequanix interface.
This pipelines requires the following executable(s):
- fastqc
- bwa
- sambamba
- samtools
- pigz
- cutadapt [or atropos]
This pipeline runs quality_control in parallel on the input fastq files (paired or not). A brief sequana summary report is also produced.
Here is the latest documented configuration file to be used with the pipeline. Each rule used in the pipeline may have a section in the configuration file.
| Version | Description |
|---|---|
| 0.11.0 |
|
| 0.10.0 |
|
| 0.9.0 |
|
| 0.8.4 |
|
| 0.8.3 |
|
| 0.8.2 |
|
| 0.8.1 | uses more sequana tools to handle the options |